Testing Within-Subjects Contrasts (Repeated Measures) in R
Within-Subjects Design
In a within-subjects design, subjects give responses across multiple conditions or across time. In other words, measures are repeated across levels of some condition or across time points. For example, subjects can report how happy they feel when they see a sequence of positive pictures and another sequence of negative pictures. In this case, we’d observe each subjects’ happiness in both positive and negative conditions. As another example, we could measure subjects’ job satisifcation every month for 3 months. Here we’d observe job satisfaction for each subject across time. In both cases, subject scores across conditions or time very likely correlate with eachother – they are dependent.
What are contrasts?
Broadly, contrasts test focused research hypotheses. A contrast comprises a set of weights or numeric values that represent some comparison. For example, when comparing two experimental group means (i.e., control vs. treatment), you can apply weights to each group mean and then sum them up. This is the same thing as subtracting one group’s mean from the other’s.
Here’s a quick demonstration
# group means
control <- 5
treatment <- 3
# apply contrast weights and sum up the results
sum(c(control, treatment) * c(1, -1))## [1] 2Model and Conceptual Assumptions for Linear Regression
- Correct functional form. Your model variables share linear relationships.
- No omitted influences. This one is hard: Your model accounts for all relevant influences on the variables included. All models are wrong, but how wrong is yours?
- Accurate measurement. Your measurements are valid and reliable. Note that unreliable measures can’t be valid, and reliable measures don’t necessairly measure just one construct or even your construct.
- Well-behaved residuals. Residuals (i.e., prediction errors) aren’t correlated with predictor variables or eachother, and residuals have constant variance across values of your predictor variables.
Model and Conceptual Assumptions for Repeated Measures ANOVA
- All change scores variances are equal. Similar to the homogenous group variance assumption in between-subjects ANOVA designs, within-subjects designs require that all change score variances (e.g., subject changes between time points, within-subject differences between conditions) are equal. This means that if you compute the within-subject differences between all pairiwse levels (e.g., timepoints, treatment levels), the variances of those parwises differences must all be equal. For example, if you ask people how satisifed they are with their current job every month for 3 months, then the variance of month 2 - month 1 should equal the variance of month 3 - month 2 and the variance of month 3 - month 1. As you might be thinking, this assumption is very strict and probably not realistic in many cases.
A different take on the homogenous change score variance assumption
- Sphericity and a special case, compound symmetry. Sphericity is the matrix algebra equivalent to the homogenous change score variance assumption. Compound symmtry is a special case of sphericity. A variance-covariance matrix that satisifies compound symmetry has equal variances (the values in the diagonal) and equal covariances (the values above or below the diagonal).
Short explanation: Sphericity = homogenous change score variance = compound symmetry
Print an example of a matrix that satisfies compound symmetry
# 3 x 3 matrix, all values = 0.20
covmat <- matrix(0.20, nrow = 3, ncol = 3)
# for kicks, make all the variances = 0.50
diag(covmat) <- rep(0.50, 3)
# print
covmat## [,1] [,2] [,3]
## [1,] 0.5 0.2 0.2
## [2,] 0.2 0.5 0.2
## [3,] 0.2 0.2 0.5Libraries
library(tidyverse)
library(knitr)
library(AMCP)
library(MASS)
library(afex)
library(lsmeans)
# select from dplyr
select <- dplyr::select
recode <- dplyr::recodeLoad data
From Chapter 11, Table 5 in Maxwell, Delaney, & Kelley (2018)
Fromhelp("C11T5"): “The data show that 12 participants have been observed in each of 4 conditions. To make the example easier to discuss, let’s suppose that the 12 subjects are children who have been observed at 30, 36, 42, and 48 months of age. Essentially, for the present data set, 12 children were each observed four times over an 18 month period. The dependent variable is the age-normed general cognitive score on the McCarthy Scales of Children’s Abilities. Interest is to determine if the children were sampled from a population where growth in cognitive ability is more rapid or less rapid than average.”
data("C11T5")
C11T5$id <- factor(1:12)
# print entire dataset
C11T5 %>% kable()| Months30 | Months36 | Months42 | Months48 | id |
|---|---|---|---|---|
| 108 | 96 | 110 | 122 | 1 |
| 103 | 117 | 127 | 133 | 2 |
| 96 | 107 | 106 | 107 | 3 |
| 84 | 85 | 92 | 99 | 4 |
| 118 | 125 | 125 | 116 | 5 |
| 110 | 107 | 96 | 91 | 6 |
| 129 | 128 | 123 | 128 | 7 |
| 90 | 84 | 101 | 113 | 8 |
| 84 | 104 | 100 | 88 | 9 |
| 96 | 100 | 103 | 105 | 10 |
| 105 | 114 | 105 | 112 | 11 |
| 113 | 117 | 132 | 130 | 12 |
Restructure data
lC11T5 <- C11T5 %>%
gather(key = month, value = score, -id) %>%
mutate(monthnum = month %>% recode("Months30" = 0, "Months36" = 6, "Months42" = 12, "Months48" = 18),
linear = monthnum %>% recode(`0` = -3, `6` = -1, `12` = 1, `18` = 3),
quadratic = monthnum %>% recode(`0` = 1, `6` = -1, `12` = -1, `18` = 1),
cubic = monthnum %>% recode(`0` = -1, `6` = 3, `12` = -3, `18` = 1),
monthfac = month %>% factor(ordered = TRUE))
# print first and last 5 observations from dataset
head(lC11T5) %>% kable()| id | month | score | monthnum | linear | quadratic | cubic | monthfac |
|---|---|---|---|---|---|---|---|
| 1 | Months30 | 108 | 0 | -3 | 1 | -1 | Months30 |
| 2 | Months30 | 103 | 0 | -3 | 1 | -1 | Months30 |
| 3 | Months30 | 96 | 0 | -3 | 1 | -1 | Months30 |
| 4 | Months30 | 84 | 0 | -3 | 1 | -1 | Months30 |
| 5 | Months30 | 118 | 0 | -3 | 1 | -1 | Months30 |
| 6 | Months30 | 110 | 0 | -3 | 1 | -1 | Months30 |
tail(lC11T5) %>% kable()| id | month | score | monthnum | linear | quadratic | cubic | monthfac | |
|---|---|---|---|---|---|---|---|---|
| 43 | 7 | Months48 | 128 | 18 | 3 | 1 | 1 | Months48 |
| 44 | 8 | Months48 | 113 | 18 | 3 | 1 | 1 | Months48 |
| 45 | 9 | Months48 | 88 | 18 | 3 | 1 | 1 | Months48 |
| 46 | 10 | Months48 | 105 | 18 | 3 | 1 | 1 | Months48 |
| 47 | 11 | Months48 | 112 | 18 | 3 | 1 | 1 | Months48 |
| 48 | 12 | Months48 | 130 | 18 | 3 | 1 | 1 | Months48 |
Visualize relationships
It’s always a good idea to look at your data. Check some assumptions.
Children scores + means and 95% confidence intervals at each time point
lC11T5 %>%
ggplot(mapping = aes(x = month, y = score)) +
geom_point(position = position_jitter(0.1)) +
stat_summary(geom = "point", fun.data = mean_cl_normal, color = "red", size = 2) +
stat_summary(geom = "errorbar", fun.data = mean_cl_normal, color = "red", width = 0) +
theme_bw()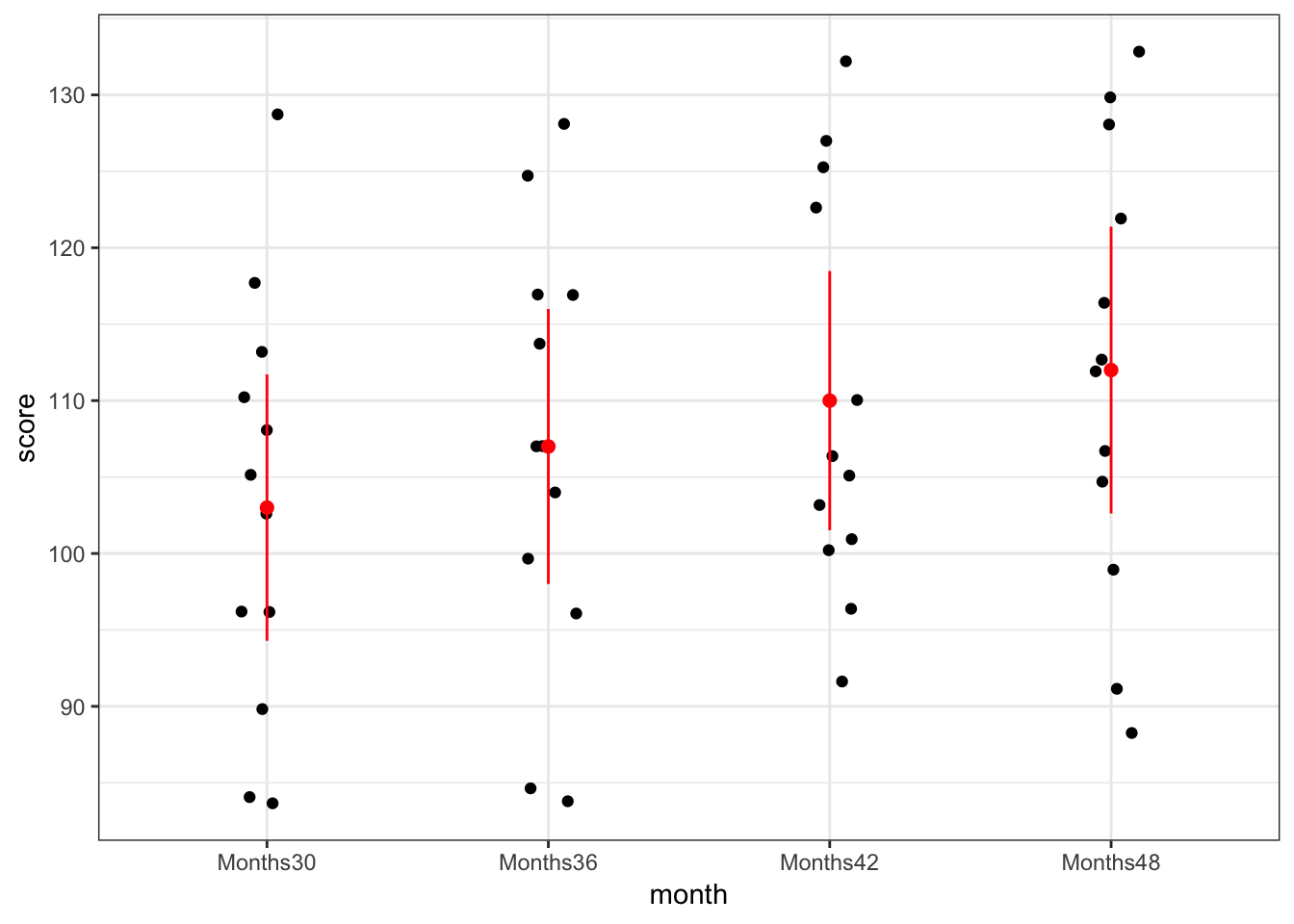
QQ-plots
Do observations look normal at each time point?
lC11T5 %>%
ggplot(mapping = aes(sample = score)) +
geom_qq() +
facet_wrap(facets = ~ month, scales = "free") +
theme_bw()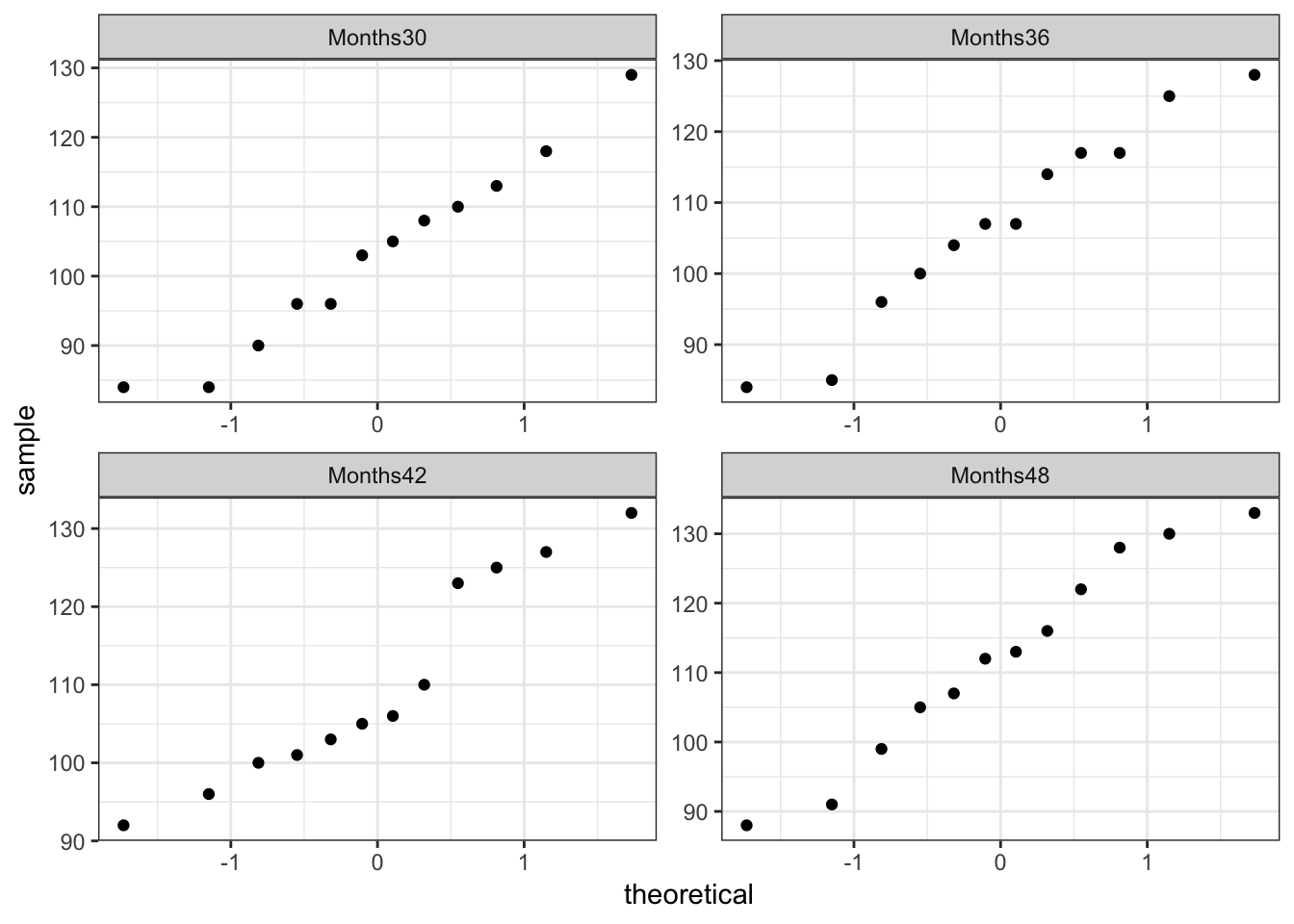
Variance-covariance matrix among 4 timepoints
Does this look like compound symmetry?
C11T5 %>% select(-id) %>% cov(use = "pairwise.complete.obs") %>% kable()| Months30 | Months36 | Months42 | Months48 | |
|---|---|---|---|---|
| Months30 | 188.0000 | 154.36364 | 127.3636 | 121.18182 |
| Months36 | 154.3636 | 200.54545 | 143.6364 | 97.45455 |
| Months42 | 127.3636 | 143.63636 | 178.0000 | 168.09091 |
| Months48 | 121.1818 | 97.45455 | 168.0909 | 218.00000 |
Test polynomial contrasts with pooled error term
Linear regression method
Below I ID use a blocking factor. This could seem nuts, but this approach “controls” for (i.e., partials out variance due to) subject ID. The polynomial contrast tests are equivalent to those you’d see from a special function or command for repeated measures ANOVA. I’ll demonstrate that later in this post.
lm01 <- lm(score ~ linear + quadratic + cubic + id, data = lC11T5)ANOVA results
anova(lm01)## Analysis of Variance Table
##
## Response: score
## Df Sum Sq Mean Sq F value Pr(>F)
## linear 1 540 540.00 8.8833 0.005369 **
## quadratic 1 12 12.00 0.1974 0.659722
## cubic 1 0 0.00 0.0000 1.000000
## id 11 6624 602.18 9.9063 1.314e-07 ***
## Residuals 33 2006 60.79
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Interpretation
We found evidence that children’s age-normed general cognitive score increased linearly over the course of 18 months, F(1, 33) = 8.88, p = .005. However, we found no sufficient evidence for a quadratic trend, F(1, 33) = 0.20, p = .660, nor a cubic trend, F(1, 33) < .001, p > .99.
Get results via algebraic formulas
Below, I test the same polynomial contrasts using algebraic equations. There’s no need to follow every last piece of code or understand each formula. I’m presenting this just to be thorough. If you’re curious, these formulas are explained in Chapter 11 of Maxwell, Dalaney, and Kelly (2018).
# contrasts
linear <- c(-3, -1, 1, 3)
quadratic <- c(1, -1, -1, 1)
cubic <- c(-1, 3, -3, 1)
# estimate (mean difference)
linest <- mean(as.matrix(C11T5[, -5]) %*% linear)
quadest <- mean(as.matrix(C11T5[, -5]) %*% quadratic)
cubest <- mean(as.matrix(C11T5[, -5]) %*% cubic)
est <- c(linest, quadest, cubest)
# number of participants
n <- nrow(C11T5)
# sum of squares for the contrast
linssc <- n * linest^2 / sum(linear^2)
quadssc <- n * quadest^2 / sum(quadratic^2)
cubssc <- n * cubest^2 / sum(cubic^2)
ssc <- c(linssc, quadssc, cubssc)
# mean squared error
mse <- anova(lm(score ~ linear + quadratic + cubic + id, data = lC11T5))$`Mean Sq`[5]
# F-ratio
fratio <- ssc / mse
# number of levels (i.e., timepoints)
nlvls <- nlevels(lC11T5$monthfac)
# degrees of freedom
df1 <- 1
df2 <- (nlvls - 1) * (n - 1)
# p-value
p <- 1 - pf(q = fratio, df1 = df1, df2 = df2)
# alpha
alpha <- 0.05
# F critical
fcrit <- qf(p = 1 - alpha, df1 = df1, df2 = df2)
# standard error
linse <- sqrt(mse * sum(linear^2 / n))
quadse <- sqrt(mse * sum(quadratic^2 / n))
cubse <- sqrt(mse * sum(cubic^2 / n))
se <- c(linse, quadse, cubse)
# lower and upper bound
lwr <- c(linest, quadest, cubest) - sqrt(fcrit) * se
upr <- c(linest, quadest, cubest) + sqrt(fcrit) * se
# table of results
tibble(est, se, fratio, p, lwr, upr)## # A tibble: 3 x 6
## est se fratio p lwr upr
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 30 10.1 8.88 0.00537 9.52 50.5
## 2 -2 4.50 0.197 0.660 -11.2 7.16
## 3 0 10.1 0 1 -20.5 20.5Get the same results using afex::aov_car
Fit model
aovcar1 <- aov_car(score ~ monthfac + Error(id / monthfac), data = lC11T5)
# equivalent
# aovez1 <- aov_ez(id = "id", dv = "score", data = lC11T5, within = "monthfac")
# aovlmer1 <- aov_4(score ~ monthfac + (1 + monthfac | id), data = lC11T5)Print polynomial contrasts
aovcar1 %>% lsmeans(specs = ~ monthfac) %>% contrast(method = "poly")## contrast estimate SE df t.ratio p.value
## linear 30 10.1 33 2.980 0.0054
## quadratic -2 4.5 33 -0.444 0.6597
## cubic 0 10.1 33 0.000 1.0000Test polynomial contrasts with separate error term
Long way
- Subset each column of data that represents a given timepoint
- Multiply that entire column of data by the contrast weight
- Add up the columns (makes one column of data)
- Test whether the mean of those values = 0 (i.e., one-sample t-test)
t.test(C11T5[, "Months30"] * -3 + C11T5[, "Months36"] * -1 + C11T5[, "Months42"] * 1 + C11T5[, "Months48"] * 3)##
## One Sample t-test
##
## data: C11T5[, "Months30"] * -3 + C11T5[, "Months36"] * -1 + C11T5[, "Months42"] * 1 + C11T5[, "Months48"] * 3
## t = 2.2414, df = 11, p-value = 0.04659
## alternative hypothesis: true mean is not equal to 0
## 95 percent confidence interval:
## 0.5403653 59.4596347
## sample estimates:
## mean of x
## 30t.test(C11T5[, "Months30"] * 1 + C11T5[, "Months36"] * -1 + C11T5[, "Months42"] * -1 + C11T5[, "Months48"] * 1)##
## One Sample t-test
##
## data: C11T5[, "Months30"] * 1 + C11T5[, "Months36"] * -1 + C11T5[, "Months42"] * -1 + C11T5[, "Months48"] * 1
## t = -0.46749, df = 11, p-value = 0.6493
## alternative hypothesis: true mean is not equal to 0
## 95 percent confidence interval:
## -11.416264 7.416264
## sample estimates:
## mean of x
## -2t.test(C11T5[, "Months30"] * -1 + C11T5[, "Months36"] * 3 + C11T5[, "Months42"] * -3 + C11T5[, "Months48"] * 1)##
## One Sample t-test
##
## data: C11T5[, "Months30"] * -1 + C11T5[, "Months36"] * 3 + C11T5[, "Months42"] * -3 + C11T5[, "Months48"] * 1
## t = 0, df = 11, p-value = 1
## alternative hypothesis: true mean is not equal to 0
## 95 percent confidence interval:
## -12.69584 12.69584
## sample estimates:
## mean of x
## 0Short way
%*%conducts a dot product of the two matrices (i.e., a time point and a contrast). This means it sums up the products of the corresponding entries of the two sequences of numbers. This is equivalent to the method above: Apply contrast weights to each column and add up the results.
t.test(as.matrix(C11T5[, -5]) %*% linear)##
## One Sample t-test
##
## data: as.matrix(C11T5[, -5]) %*% linear
## t = 2.2414, df = 11, p-value = 0.04659
## alternative hypothesis: true mean is not equal to 0
## 95 percent confidence interval:
## 0.5403653 59.4596347
## sample estimates:
## mean of x
## 30t.test(as.matrix(C11T5[, -5]) %*% quadratic)##
## One Sample t-test
##
## data: as.matrix(C11T5[, -5]) %*% quadratic
## t = -0.46749, df = 11, p-value = 0.6493
## alternative hypothesis: true mean is not equal to 0
## 95 percent confidence interval:
## -11.416264 7.416264
## sample estimates:
## mean of x
## -2t.test(as.matrix(C11T5[, -5]) %*% cubic)##
## One Sample t-test
##
## data: as.matrix(C11T5[, -5]) %*% cubic
## t = 0, df = 11, p-value = 1
## alternative hypothesis: true mean is not equal to 0
## 95 percent confidence interval:
## -12.69584 12.69584
## sample estimates:
## mean of x
## 0Custom function for testing via both methods
This function takes “wide-form” data and a contrast as input and it outputs a matrix with two columns: p-values and which testing approach they came from, pooled or separate error term.
wscont <- function(wdata, cont) {
# long data
ldata <- stack(wdata)
colnames(ldata) <- c("y", "t")
# add id factor; make time a factor
ldata$id <- factor(rep(1:nrow(wdata), times = ncol(wdata)))
ldata$t <- factor(ldata$t, ordered = TRUE)
# separate variance contrast p-value
svp <- t.test(as.matrix(wdata) %*% cont)$p.value
# fit model
fit <- lm(y ~ t + id, data = ldata)
# extract p-value from linear contrast
pvp <- summary.aov(fit, split = list(t = list(lin = 1)))[[1]][, "Pr(>F)"]["lin"]
# output
cbind(sepvar = c(0, 1), p = c(pvp, svp))
}Simulate false positive error rates that result from both approaches
Maxwell, Dalaney, and Kelly (2018) refer to these approaches as the univariate approach and the multivariate approach. To make the labels more descriptive, I refer to each approach by the error term it uses: pooled vs. seperate (MDK2018 do too)
True Covariance Matrix
Here’s an example of a covariance matrix that satisifes the homogenous change score variance assumption. All timepoint variances are equal and so are the covariances between timepoints = 0. This is also called an identitiy matrix.
(truecov <- diag(1, 4, 4))## [,1] [,2] [,3] [,4]
## [1,] 1 0 0 0
## [2,] 0 1 0 0
## [3,] 0 0 1 0
## [4,] 0 0 0 1Sample data
Below I generate data that represent 10 subjects’ scores across 4 times points. The mean score at each timepoint is 0, and the correlations among timepoints stem from the true matrix I saved above.
# set random seed so results can be reproduced
set.seed(8888)
dat <- data.frame(mvrnorm(n = 10, mu = c(0, 0, 0, 0), Sigma = truecov))Sample test
Here I test out the function I made above.
wscont(wdata = dat, cont = linear)## sepvar p
## lin 0 0.12806078
## 1 0.04189252Generate data that satisfy compound symmetry
Compound Symmetric Covariance Matrix
(cscovmat <- diag(0.5, nrow = 4, ncol = 4) + 0.5)## [,1] [,2] [,3] [,4]
## [1,] 1.0 0.5 0.5 0.5
## [2,] 0.5 1.0 0.5 0.5
## [3,] 0.5 0.5 1.0 0.5
## [4,] 0.5 0.5 0.5 1.0Save true means over time
truemeans <- c(100, 100, 100, 100)Data
- Create a function returns a data.frame with the requested means and variance-covariance structure (compound symmetry in this case)
- Save a data.frame with 10,000 n = 50 replications
# function
gendatafun <- function(truecov, ms, n) {
data.frame(mvrnorm(n = n, mu = ms, Sigma = truecov))
}
# data generation
simdata1 <- do.call(rbind, replicate(n = 10000, gendatafun(truecov = cscovmat, ms = truemeans, n = 50), simplify = FALSE))Generate data with the same variance-covariance structure as example data
Save covariance matrix from data used in Chapter 11, Table 5 of Maxwell, Dalaney, and Kelley (2018)
(c11t5covmat <- cov(C11T5[, -5]))## Months30 Months36 Months42 Months48
## Months30 188.0000 154.36364 127.3636 121.18182
## Months36 154.3636 200.54545 143.6364 97.45455
## Months42 127.3636 143.63636 178.0000 168.09091
## Months48 121.1818 97.45455 168.0909 218.00000Data
These data have a variance-covariance structure like the Chapter 11, Table 5 data.
# data generation
simdata2 <- do.call(rbind, replicate(n = 10000, gendatafun(truecov = c11t5covmat, ms = truemeans, n = 50), simplify = FALSE))Add replicate IDs
I’ll use this column later to subset each replication.
simdata1$i <- rep(1:10000, each = 50)
simdata2$i <- rep(1:10000, each = 50)Get p-values via custom wscont() function
In this last line, I add a column, compsymm, that flags which population variance-covariance matrix the data came from (0 = C11T5 matrix, 1 = compound symmetry).
simp1 <- 1:10000 %>% map_df(function(rep) {
simdata1 %>%
filter(i == rep) %>%
select(-5) %>%
wscont(cont = linear) %>%
as_tibble() %>%
mutate(compsymm = 1)
})
simp2 <- 1:10000 %>% map_df(function(rep) {
simdata2 %>%
filter(i == rep) %>%
select(-5) %>%
wscont(cont = linear) %>%
as_tibble() %>%
mutate(compsymm = 0)
})Plot false positive errors
p < .05 should only occur 5% of the time
bind_rows(simp1, simp2) %>%
mutate(sig = ifelse(p < .05, 1, 0)) %>%
group_by(compsymm, sepvar) %>%
summarise(falsepos = mean(sig)) %>%
ungroup() %>%
mutate(sepvar = sepvar %>% recode(`0` = "Pooled Error Term", `1` = "Separate Error Term"),
compsymm = compsymm %>% recode(`0` = "Unequal Variances & Covariances", `1` = "Compound Symmetry")) %>%
ggplot(mapping = aes(x = compsymm, y = falsepos, fill = sepvar)) +
geom_bar(stat = "identity", position = position_dodge(0.9)) +
geom_hline(yintercept = 0.05, color = "red") +
scale_y_continuous(breaks = seq(0, 0.5, 0.01)) +
theme_bw() +
theme(legend.position = "top")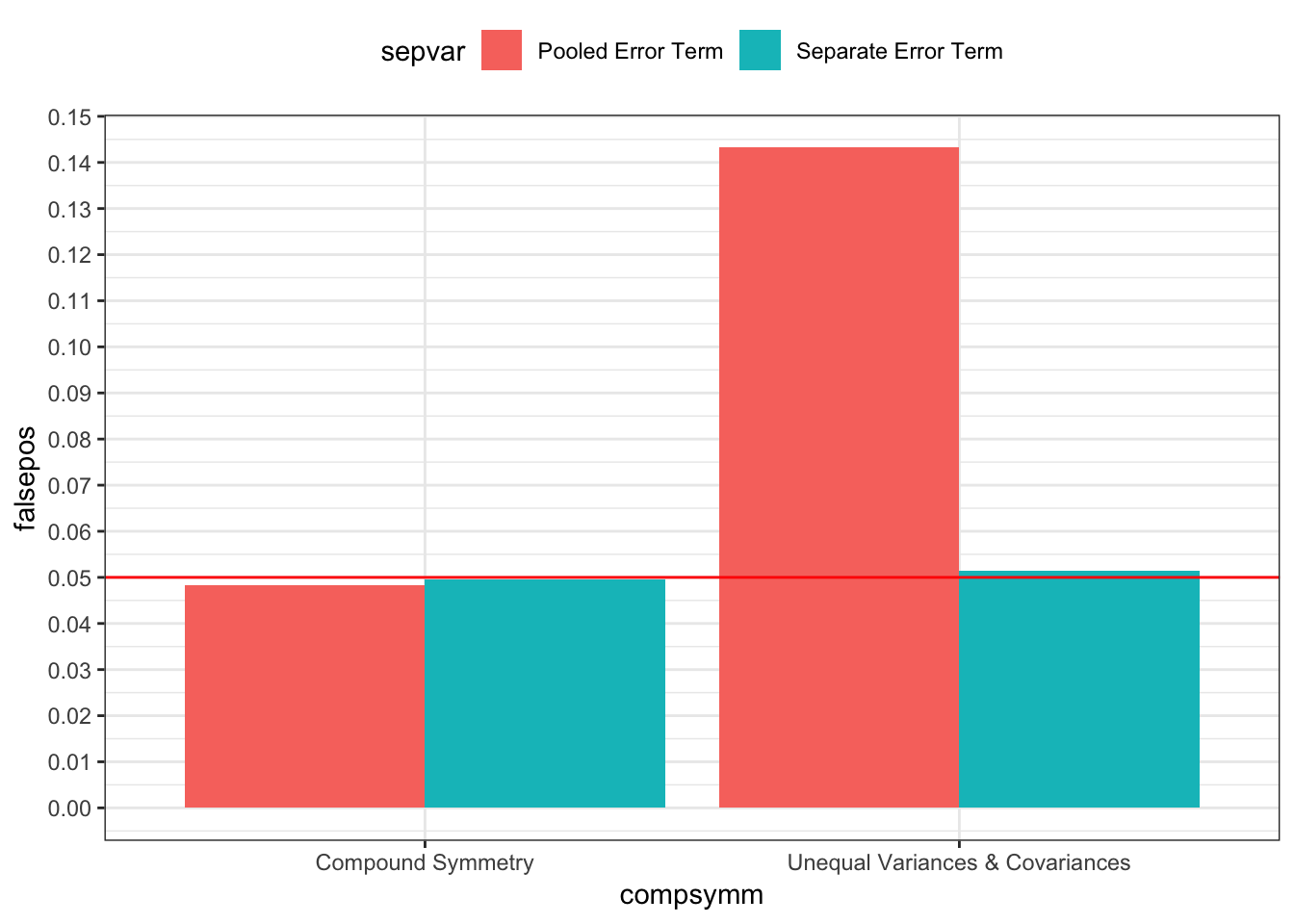
Interpretation
Above, I generated 20,000 datasets with known population variance-covariance matrices and columns means. For each dataset, I tested the linear contrast across time using both error term approaches and saved the p-values. When the true population variance-covariance matrice satisfied compound symmetry, both approaches’ contrast test statistics give practically the same results, on average, but when the true population variance-covariance matrice violated compound symmetry – a very common scenario in real datasets – the pooled error term test statistics resulted in false positives over 14% of the time; the separate error term test statistics resulted in false positives only 5% of the time.
Resources
- Boik, R. J. (1981). A priori tests in repeated measures designs: Effects of nonsphericity. Psychometrika, 46(3), 241-255.
- Maxwell, S. E., Delaney, H. D., & Kelley, K. (2018). Designing experiments and analyzing data: A model comparison perspective (3rd ed.). Routledge.
- DESIGNING EXPERIMENTS AND ANALYZING DATA includes computer code for analyses from their book, Shiny apps, and more.
General word of caution
Above, I listed resources prepared by experts on these and related topics. Although I generally do my best to write accurate posts, don’t assume my posts are 100% accurate or that they apply to your data or research questions. Trust statistics and methodology experts, not blog posts.